Now let's look at the combinatorial approach to store molecules: We can fit a Chemical Space containing 290,000,000,000,000 (2.9 x 10
14) molecules into 13.5 MB; this very space is also known as the Knowledge Space.
How is this possible?
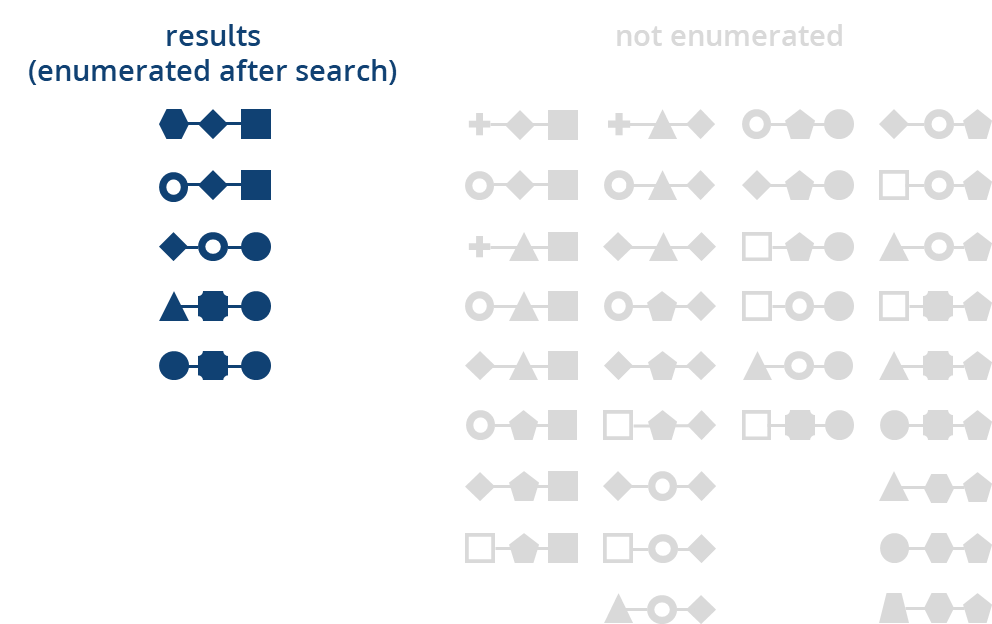
The Knowledge Space has been created using a diverse set of molecular building blocks and robust chemical reactions that can be performed in most synthesis labs by almost everyone with a background in chemistry. So instead of storing all the compounds, we store the recipe to make them. Searching this space with BioSolveIT applications can yield every possible entry of the 2.9 x 10
14 molecules as a hit — by applying the reaction rules how to combine one building block with another.
It is possible to create your own Chemical Space with one of BioSolveIT's Amazing Workflows.