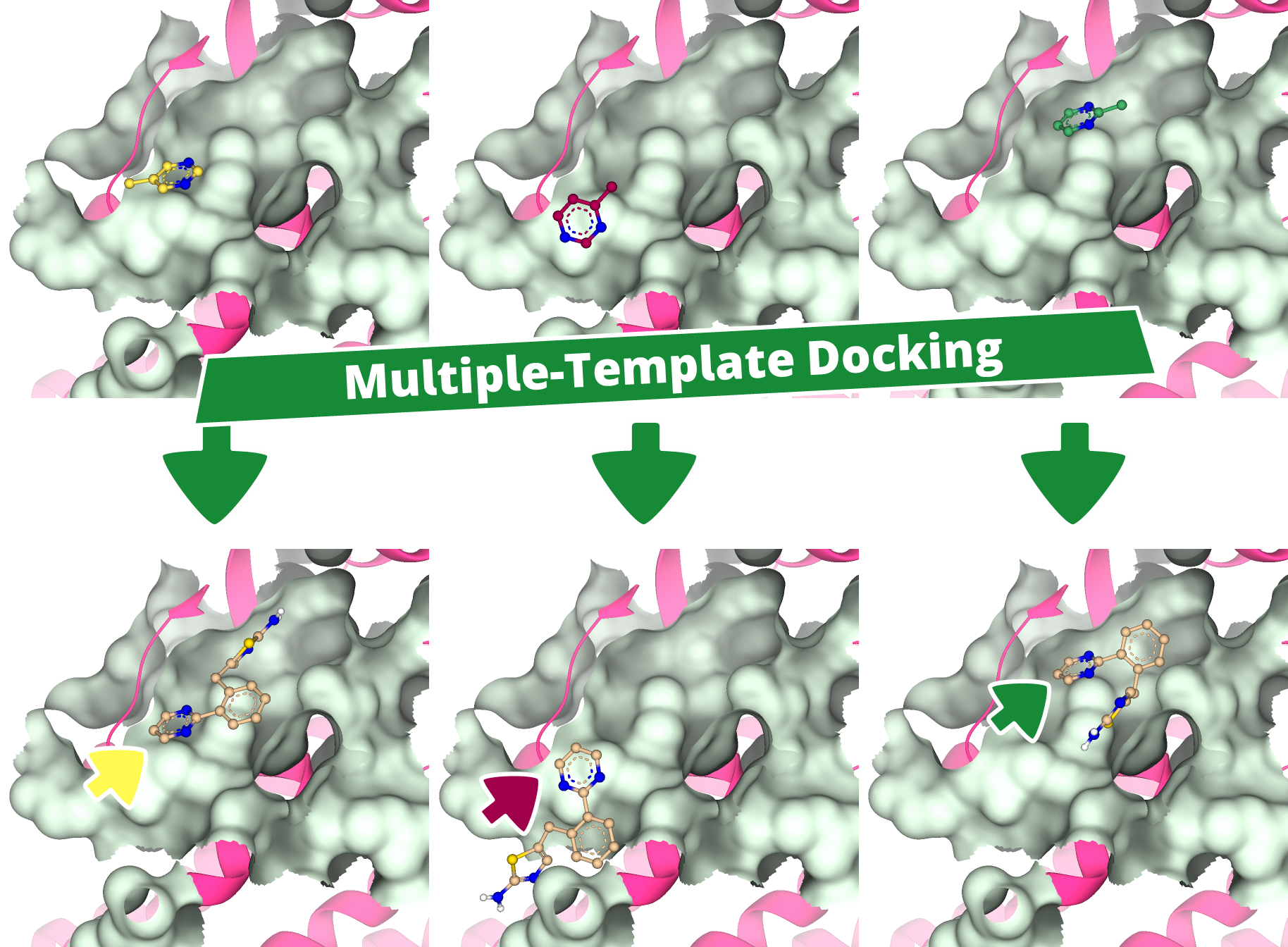
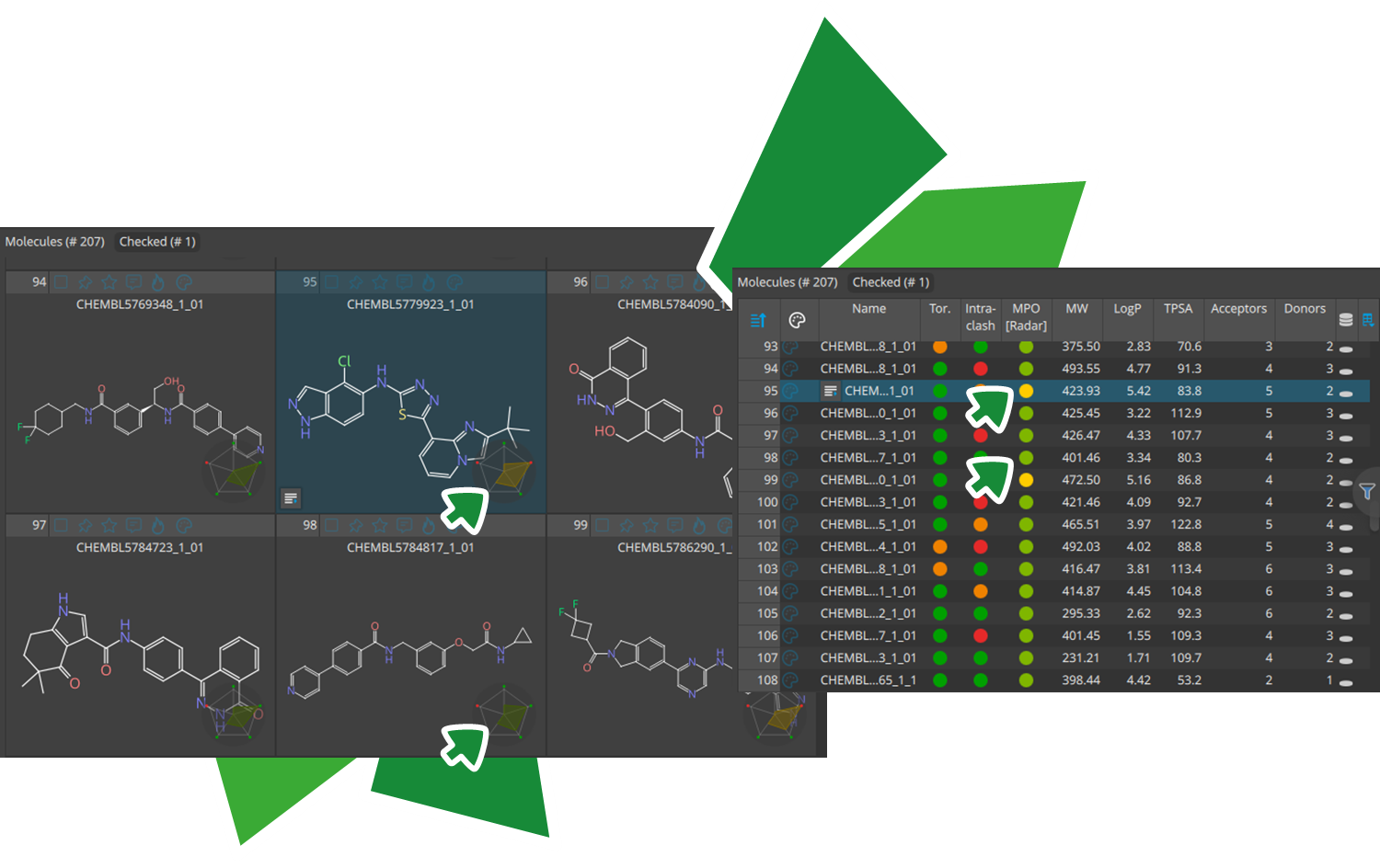
Introducing 'Apollo'
We are excited to share a preview of the upcoming drug design dashboard SeeSAR 15 release, proudly titled 'Apollo.'
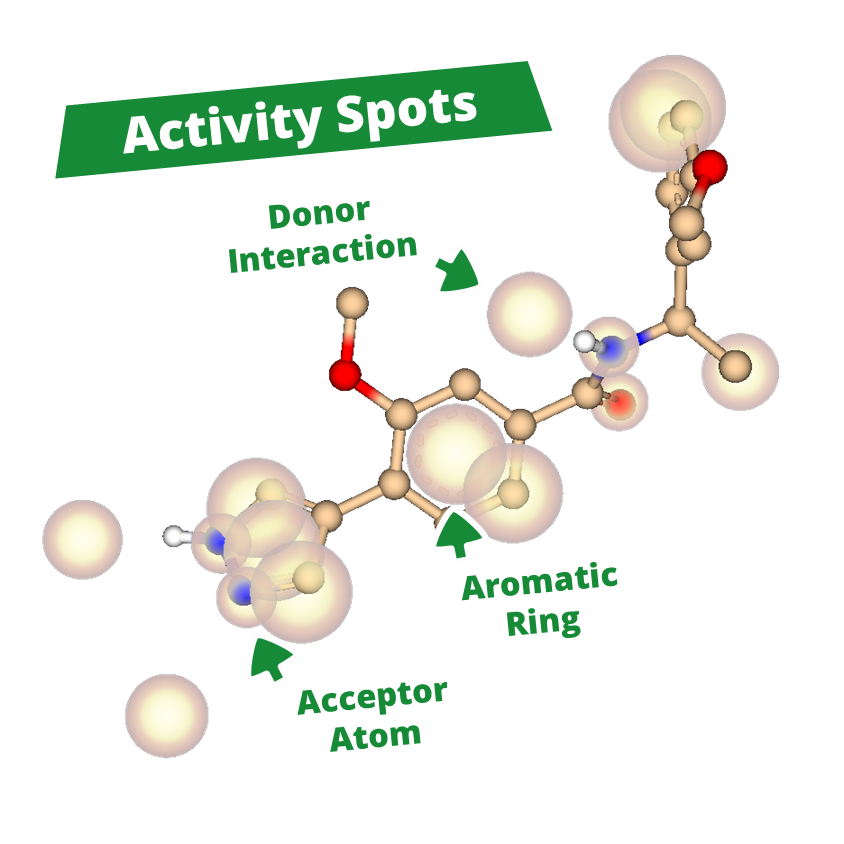
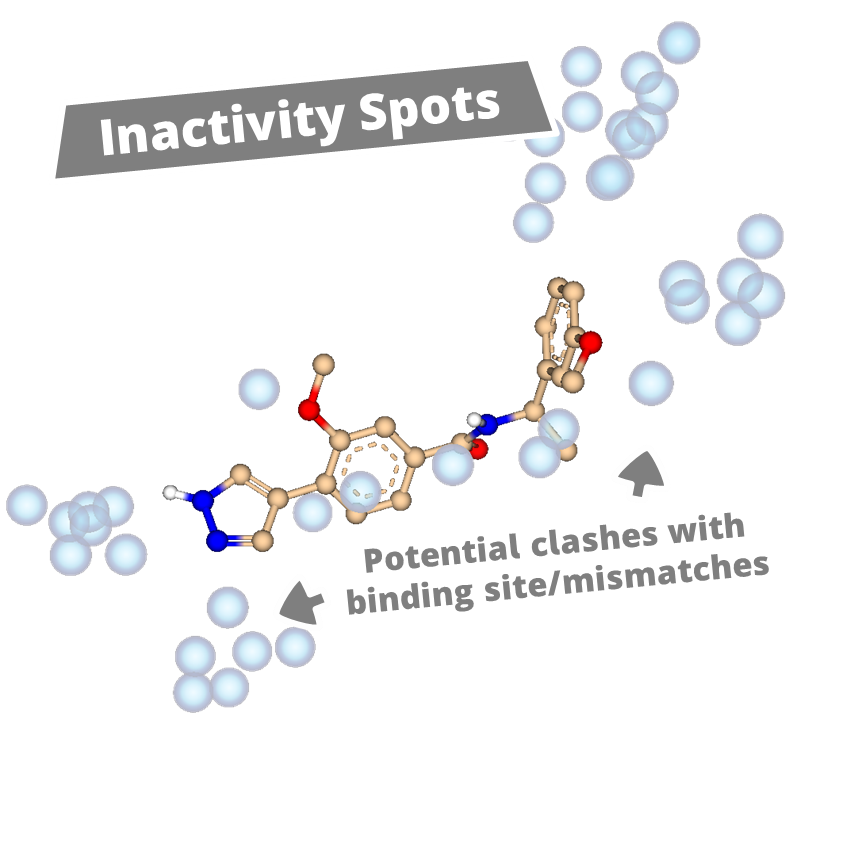
With powerful new features, Apollo enables users to uncover new insights and develop a clearer view of a compound series. The new Activity Spotter Mode, analyzes compound activity to clarify structure-activity relationships and turn findings into actionable guidance for optimization and screening.
Here, we present the highlights of the upcoming SeeSAR release.