- Now in your BioSolveIT toolkit: The Synple Space is officially accessible across BioSolveIT platforms and tools.
- Fast ligand-based screening: Multiple similarity search methods let you retrieve accessible compounds and assemble custom, project-specific libraries in moments.
- Local and secure: Searches complete in seconds to minutes on standard hardware—no cloud or third-party dependency, no data leaving your environment.
- Structure-based virtual screening: Run virtual screening campaigns over Synple Space with Chemical Space Docking®, finishing in a fraction of the time compared to brute-force screening of trillion-sized collections.
Browsing the Ultra-sized Collection in Seconds
The Synple Chemical Space is now fully accessible across all BioSolveIT platforms, making trillion-scale exploration convenient for state-of-the-art small molecule discovery. You can launch searches on your own standard workstation and see results in seconds to minutes, keeping IP in-house while dramatically shortening the computational process.
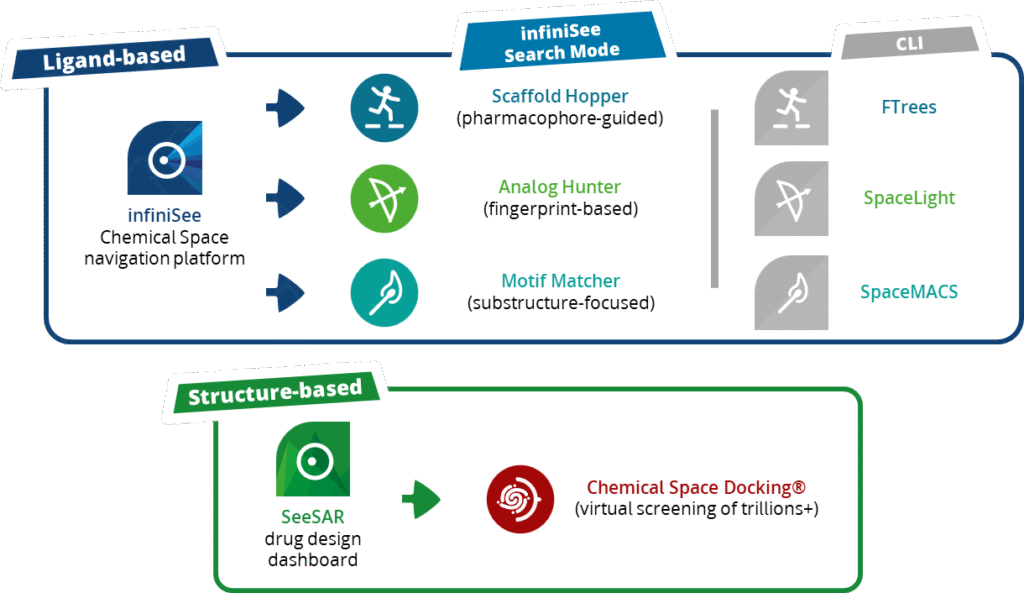
BioSolveIT’s Chemical Space navigation platform, infiniSee, lets you retrieve the most relevant chemistry for your project in a user-friendly environment. Users can select from different, orthogonal ligand-based methods, including pharmacophore-guided, molecular fingerprint-based, and substructure-focused searches, to explore different areas of the chemical spaces for molecular resemblance. These methods are also available as parameterizable command-line tools (FTrees, SpaceLight, and SpaceMACS, respectively) for computational workflows and automation, so accessible compounds can be collected and custom libraries be assembled exactly the way the project demands.
Structure-Based Screening at Trillion Scale
With Synple Space embedded across the BioSolveIT environment, the collection becomes directly usable for structure-based virtual screening via Chemical Space Docking®. During a run, the method samples the trillion-sized Synple Space and evaluates compounds for their interactions at the target’s binding site, ranking the most promising chemotypes for follow up.
Because Chemical Space Docking® samples the Space combinatorially rather than exhaustively enumerating it, it delivers decision-ready hit lists in a fraction of the time typically associated with screening collections of this magnitude. Workflows run securely on your local hardware, are fully parameterizable (e.g., pharmacophore constraints, template guidance, property filters), and hand off cleanly to BioSolveIT tools for triage and design.
How BioSolveIT and Synple Synergize
Together, BioSolveIT and Synple close the loop from design to make: Trillion-scale search meets on-demand, automated synthesis, all in a local environment built for speed and control.
- Offline and air-gapped by design: Searches run entirely on local machines. No data leaves your environment which is ideal for IP-sensitive and regulated settings.
- Parameterizable CLI: Rich, tunable command-line controls leave you in full control for the search runs to fine-tune your results.
- Much faster than standard approaches: Ligand-based retrieval returns candidates in seconds to minutes, and Chemical Space Docking® samples smater than exhaustive screening, reducing computational costs to only a fraction.
- From virtual to vial: Hits identified in the software are on-demand synthesizable on Synple instruments, enabling rapid follow-up and iteration.
How to access the Synple Space
Screening the Synple Space is straightforward and flexible. For ligand-based approaches, use BioSolveIT’s InfiniSee for an interactive, visual workflow, or plug the same capabilities into pipelines via the command-line tools FTrees, SpaceLight, and SpaceMACS.
For structure-based campaigns, Chemical Space Docking® is available in BioSolveIT’s drug-design dashboard SeeSAR.





