Molecular superpositioning of small molecules is one of the pillars of ligand-based drug discovery (LBDD). In LBDD cheminformatics methods are applied to 3D align a molecule set with a query structure based on tool criteria, e.g., motifs, interaction features, and substructures. Subsequently, the 3D alignment can be exploited to investigate SAR (structure-activity relationships), pharmacophore feature generation, or — using an alignment or superposition score — simply as a virtual screening method.
The latter is of great interest for drug discovery projects as it can be applied in cases where no target structure is available, or the target is not known at all. Each member of a library of compounds is superposed onto the query, and the associated scoring is computed. In other words, the method uses a scoring function to determine the “similarity” of the 3D alignment amongst all structures of the library, so that we can rank the results for selection of candidates for a subsequent in vitro assessment.
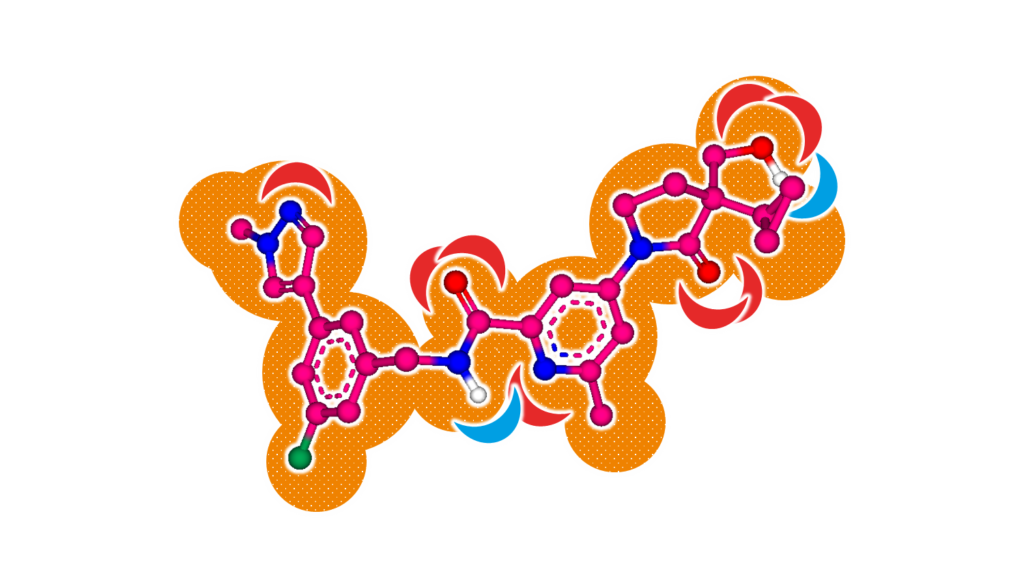
SeeSAR’s Similarity Scanner performs molecular superpositioning using ideas of the FlexS algorithm. It 3D-aligns molecules based on their interaction features (e.g. H-bond donor/acceptor), volume and shape — and it certainly also provides an alignment score (see Figure 1). Given a known active query compounds, molecule libraries can be aligned to perform virtual screening for candidates that share resemblance to the query compound. The results can be fine-tuned or filtered by applying pharmacophore constraints.
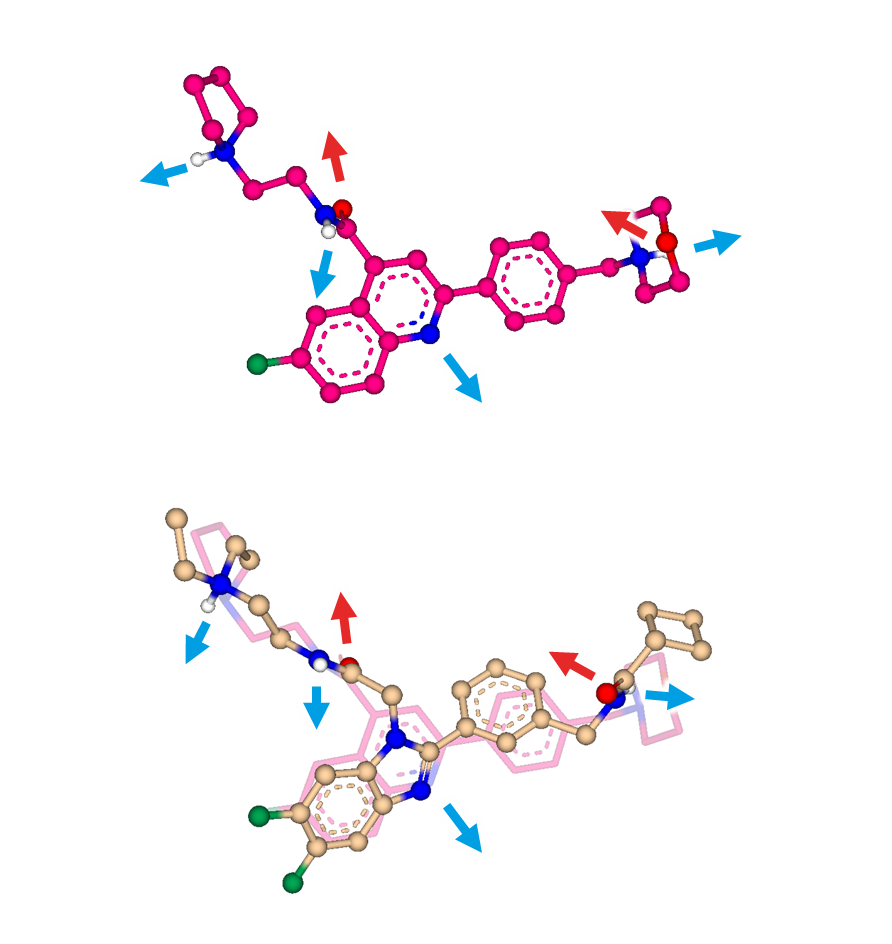
Figure 1. Molecular superposition performed by SeeSAR’s Similarity Scanner and the FlexS algorithm. Blue arrows indicate interaction points for H-bond donors. Red arrows indicate interaction points for H-bond acceptors.
The Similarity Scanner is a great enrichment method to post-process Chemical Space searches: In a case study we used a query compound (see Figure 2) to screen make-on-demand Chemical Spaces with infiniSee for close analogs. Afterwards, the results were loaded into SeeSAR for 3D alignment with the query compound, and finally they were ranked. The results displayed broad chemical diversity among the spaces originating from the in-house knowledge and building blocks. Comparison of the results with the query compound revealed the potential for scaffold hopping and provided suggestions for other functional groups as replacement. The leap into the 3D world provides power for the discovery of potential candidates which would not be accessible through 2D structure alignment or Tanimoto similarity screening alone.
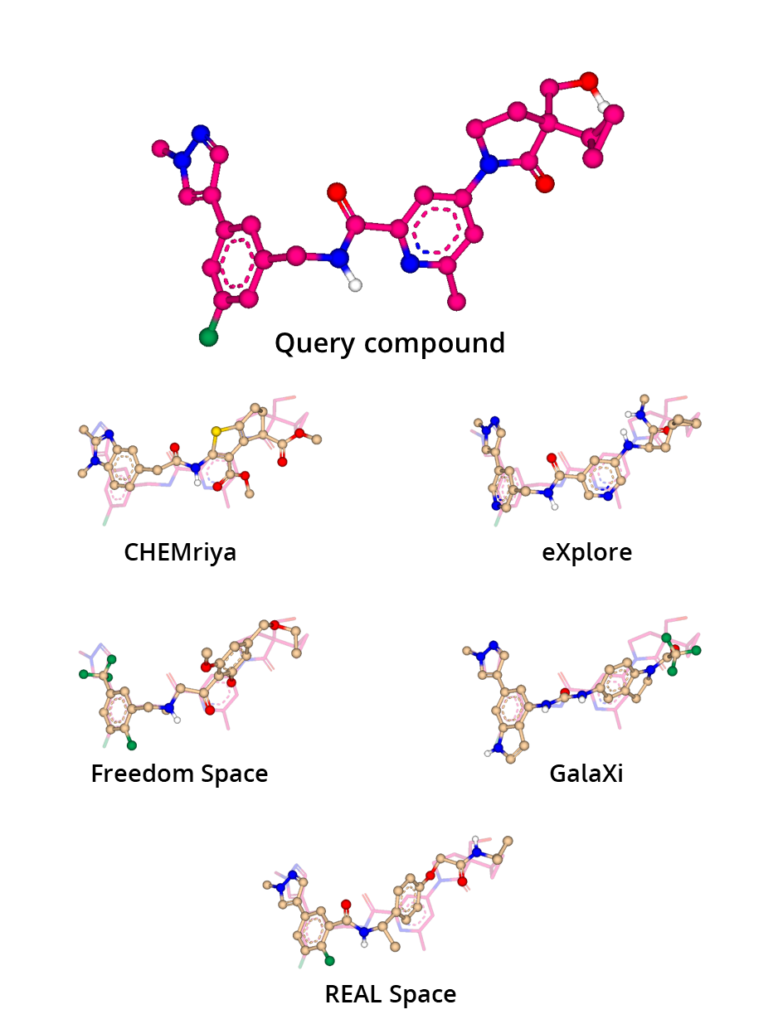 Figure 2. Molecular superposition of molecules retrieved by infiniSee from Chemical Spaces with a query compound.
Figure 2. Molecular superposition of molecules retrieved by infiniSee from Chemical Spaces with a query compound.
The Similarity Scanner is part of BioSolveIT’s drug discovery dashboard SeeSAR. The 3D alignment tool is also available as a stand-alone command line tool. You can discover more examples and use-cases of the Similarity scanner in this recording (Youtube) of one of our workshops.
If you want to learn more about molecular superpositioning: An new, insightful and broad review can be accessed here.